Quantitative Polymerase Chain Reaction (a.k.a qPCR) is the way of detecting the amount of product obtained in the Polymerase Chain Reaction. Detecting the presence and measuring the amount of mRNA transcript in a particular part of an organism at a particular time is one of its best applications known as expression profiling of genes.
Right primers are one of the most sensitive requirements of the Good Polymerase Chain Reaction (PCR). PCR can fail if the Primers designed before the reaction are not the right ones. There are several Online and desktop-based Primer designing software available but no one can give you the right pair of primers without their proper knowledge in advance. In this guide, we will design specific primers using the NCBI Primer-BLAST online tool.
Step 1: You will need the FASTA sequence, accession or GI of your target portion which you want to amplify. Becuase we have assumed that we are working with expression profiling, we must use CDS of the gene. If you don’t have the CDS yet, you can predict CDS using ORF. If you just want to amplify that particular DNA sequence than the Genomic FASTA will also work.
Step 2: Fill the form while keeping the following things in mind.
- For qPCR, keep the Product size 200-300
- Select the particular organism such as Homo sapiens or Zea mays
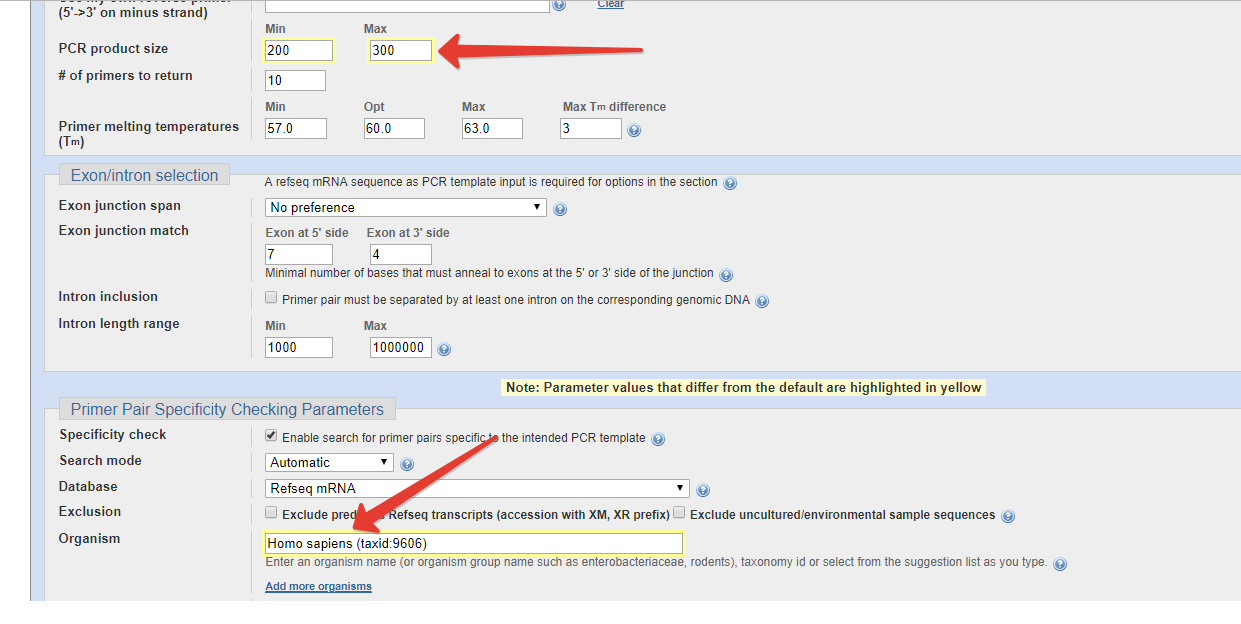
To make that the inserted FASTA has no similar presence in rest of the genome of that particular organism, the tool will first BLAST the whole genome for that sequence and if it finds one, it will show. Normally it is the sequence of your gene.
Step 3: The results page will show up to 10 results ranging in the different regions of the submitted sequences. Coincidently in the current case, all the primers are lying in the same region.
You may choose the primers pair using following information,
- GC ratio must be around 50
- Optimum length of primer 18-20
- Difference between the Tm around 1 ° C
- Product length 200-300 (Which we already defined)
- Less GCs at the end
Want to suggest any Edit or ask any Question? Feel free to reply in the comment box